This research explores 25 years of curated genetic data to identify critical genomic regions linked to key traits in Prunus crops such as peaches, almonds, apricots, and sweet cherries.
By analyzing over 28,000 trait-associated loci from genome-wide studies, the researchers have pinpointed QTL hotspots that regulate fruit quality, morphology, phenology, and disease resistance.
These findings aim to improve Prunus breeding programs, offering actionable data that can enhance the genetic improvement of economically significant crops.

Genetic insights for breeding
This work also highlights the potential of using syntenic regions across different species to streamline breeding efforts, especially for polyploid genomes and lesser-studied species.
Prunus species, including peaches, cherries, and almonds, are vital to global agriculture, with millions of tons produced annually.
Traditional breeding techniques have relied on phenotypic selection and basic genetic markers.
However, modern molecular technologies now provide deeper insights into the genetic basis of crucial traits.
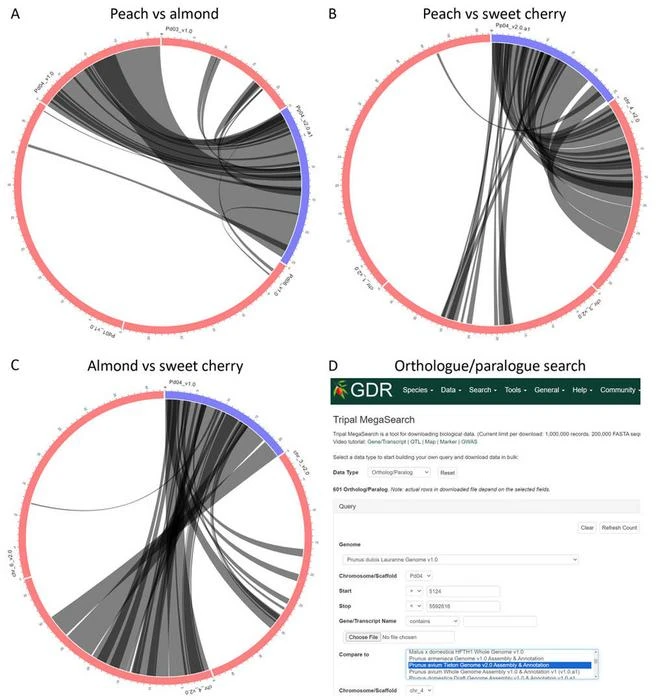 Image 1. Chr 4 syntenic relationships in Prunus. Synteny between (A) peach and almond, (B) peach and sweet cherry, (C) almond and sweet cherry, and (D) an example of orthologue/paralogue search using the genome positions obtained from the synteny analysis of almond versus sweet cherry. Blue lines represent selected chromosomes; red lines are chromosomes from the genome being compared to; black lines are syntenic blocks.
Image 1. Chr 4 syntenic relationships in Prunus. Synteny between (A) peach and almond, (B) peach and sweet cherry, (C) almond and sweet cherry, and (D) an example of orthologue/paralogue search using the genome positions obtained from the synteny analysis of almond versus sweet cherry. Blue lines represent selected chromosomes; red lines are chromosomes from the genome being compared to; black lines are syntenic blocks.
Role of genomic databases
The Genome Database for Rosaceae (GDR) has played a pivotal role in curating genetic and genomic data, enabling researchers to map key traits across Prunus species.
These advances have been driven by high-throughput genotyping and sequencing technologies, which have revolutionized the way breeders approach crop improvement.
Based on these challenges, there is a need to integrate genetic data across species for better breeding strategies.
Study findings and hotspots
Researchers from Clemson University and Washington State University recently published (DOI: 10.1093/hr/uhaf142) their findings in Horticulture Research (May 2025).
The study analyzes curated data from the Genome Database for Rosaceae (GDR) covering 25 years of genetic research.
The study identifies major quantitative trait loci (QTLs), Mendelian trait loci (MTLs), and genome-wide association study (GWAS) results, offering new insights into the genetic architecture underlying crucial traits in Prunus species.
This work provides breeders with valuable resources for enhancing crop yield, disease resistance, and fruit quality.
QTL hotspots and synteny
The research focuses on genetic loci associated with key traits in Prunus species, using data from 177 genetic maps and 28,971 trait-associated loci, including QTLs, MTLs, and QTNs.
The study emphasizes the identification of 16 QTL hotspots linked to traits like fruit quality, growth phenology, and disease resistance.
Notably, 76.4% of the loci are related to morphological and quality traits, reflecting the breeding community's focus on consumer preferences.
The study also reveals the genetic collinearity across species, identifying 17 syntenic regions among peach, sweet cherry, and almond genomes.
Applications for breeders
These syntenic regions are invaluable for breeders, as they enable the transfer of genetic information across species and provide a roadmap for improving complex traits in polyploid Prunus species.
Furthermore, the study highlights the role of high-priority QTL hotspots, which offer a foundation for developing molecular tools that could accelerate breeding programs.
The ability to map and transfer these genetic markers across different Prunus species, especially for lesser-studied crops, could significantly advance breeding efforts, reducing the time required to develop improved cultivars.
Expert perspective and impact
Dr. Michael Itam, a lead researcher from Clemson University, commented on the importance of the findings: "Our work opens up new possibilities for breeding Prunus crops with enhanced traits such as disease resistance and improved fruit quality.
The integration of shared molecular data across different species will help breeders make more informed decisions and optimize breeding strategies, particularly for species with complex genomes like peaches and almonds.
The identification of QTL hotspots and syntenic regions will be instrumental in accelerating the development of better-performing cultivars."
This research provides a critical resource for Prunus breeding, highlighting the genetic hotspots that control economically important traits.
By utilizing these insights, breeders can more effectively select for desired traits, such as fruit quality, yield, and disease resistance, in Prunus species.
The identification of syntenic regions also offers the potential to transfer genetic information between species, speeding up the breeding process for both well-studied and under-researched species.
This approach not only enhances the efficiency of breeding programs but also promotes the sustainable production of high-quality Prunus crops, ensuring better food security and agricultural resilience in the face of changing environmental conditions.
Source: Ping Wang - Horticulture Research, Nanjing Agricultural University
Opening image source: Tasting Table
Cherry Times - All rights reserved













