As is well known, a gametophytic self-incompatibility system (GSI) determines fertility in sweet cherry varieties and prevents pollination with self and genetically related pollen. This mechanism is controlled by a multiallelic S locus that encodes two genes involved in fertilization: the allele-specific ribonuclease S (S RNase) expressed in the female style and the S-specific F-box protein (SFB) in the male pollen.
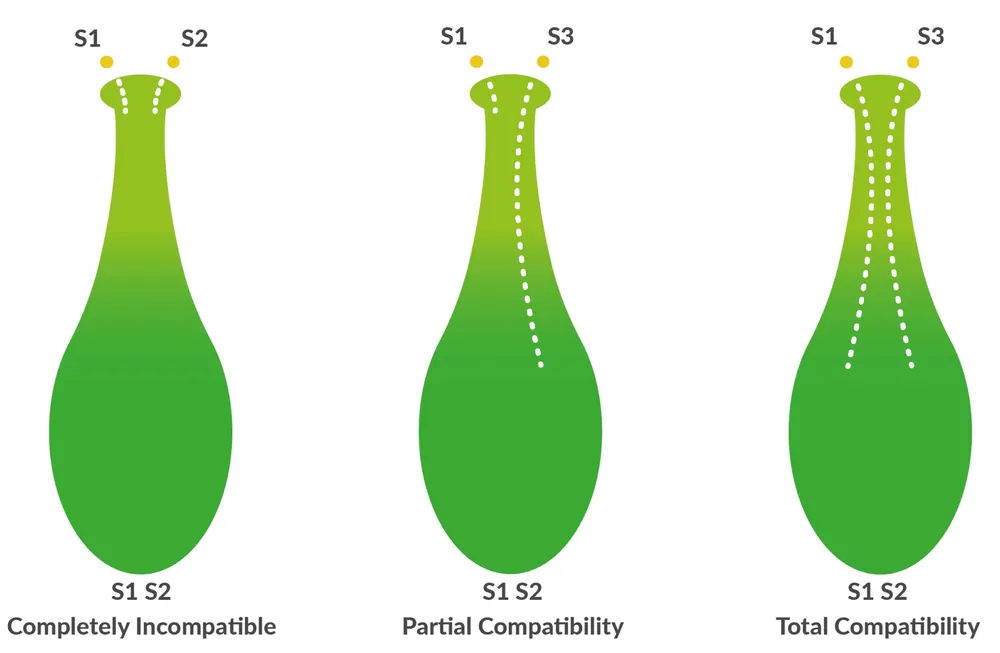 Figure 1. Graphical representation of incompatibility mechanisms in sweet cherry.
Figure 1. Graphical representation of incompatibility mechanisms in sweet cherry.
The knowledge of the S genotype of sweet cherries is very important for cherry growers. In fact, to achieve good pollination and fruit set, in addition to an adequate presence of pollinators and the correct choice of varieties with overlapping bloom, perfect genetic compatibility between the main variety and its pollinator must be ensured.
Over 1700 varieties analyzed
Over the years, molecular markers have been developed to distinguish S alleles in sweet cherries. In a recent update by Schuster and Schröpfer of the JKI - Julius Kühn-Institut (Germany), a total of 72 incompatibility groups in 1700 sweet cherry cultivars were listed.
Twenty-six sweet cherry cultivars have a unique combination of S alleles, have been described as universal donors, and were categorized into incompatibility group 0. Additionally, there are 91 sweet cherry genotypes that are self-compatible (SC) or self-fertile (SF) and belong to group SC.
So far, a total of 26 different S alleles have been described in cultivated sweet cherries (for further details: https://cherrytimes.it/news/una-panoramica-sugli-s-alleli-delle-ciliegie-dolci-coltivate).
The interactive allele atlas
Building on the work of the German research group, Jesús Alonso Sánchez, a researcher at the Spanish National Research Council (CSIC), has created a kind of easily searchable online database.
To access the file click here
Cherry Times editorial staff
Image source: SL Fruit Service; Primrose
Cherry Times - All rights reserved











