The Chinese cherry, Prunus pseudocerasus, is an important crop in China with a cultivation history spanning thousands of years. However, a limiting factor for its widespread commercial distribution is the fruit’s low firmness, which reduces its ability to be transported over long distances.
Thanks to recent studies, researchers have managed to obtain a detailed analysis of the Chinese cherry genome, paving the way for new possibilities in the genetic improvement of this species and, consequently, its broader distribution.
The sequencing of the tetraploid genome of the Chinese cherry was carried out using cutting-edge sequencing technologies, such as PacBio HiFi and Oxford Nanopore, which enabled the production of a high-resolution reference genome. The main difficulty in sequencing this species lies in its tetraploid and highly heterozygous nature. With the newly haplotype-resolved genome, researchers were able to thoroughly analyze the genomic structure of the species.
Phylogenetic and genomic analyses revealed that Prunus pseudocerasus is a stable autotetraploid species, derived from a genome duplication event that occurred approximately 139.96 million years ago. By comparing the Chinese cherry genome with those of other Prunus species, such as sweet cherry (Prunus avium) and wild cherry (Prunus pusilliflora), it was observed that the two species diverged around 18.34 million years ago.
However, one of the main differences between these species concerns fruit texture: Prunus pseudocerasus has much softer fruits compared to sweet cherry.
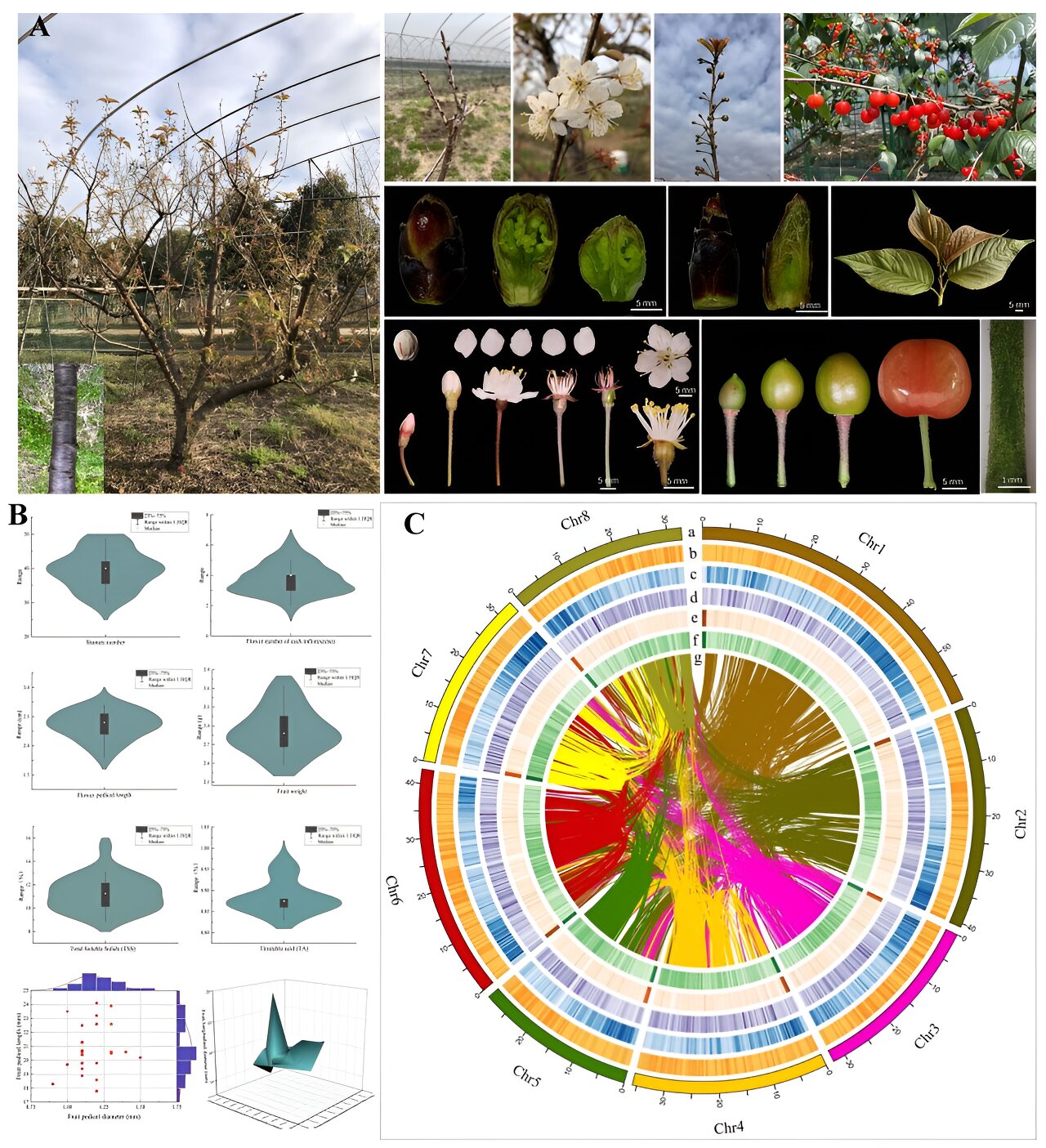 Image 1. Source: Songtao Jou.
Image 1. Source: Songtao Jou.
The study demonstrated that this difference in texture is linked to the composition of the cell wall. In the case of the Chinese cherry, the fruit's cell wall contains lower amounts of cellulose, hemicellulose, and pectin compared to sweet cherry, especially during the ripening phase. This difference was linked to the activity of specific genes involved in pectin synthesis, particularly “GalAK-like” and “Stv1,” two key genes identified in the comparative analysis between the Chinese and sweet cherry genomes.
Researchers discovered that the overexpression of these two genes leads to a significant increase in fruit firmness. Specifically, the increased activity of GalAK-like and Stv1 results in an accumulation of protopectin, a precursor of pectin. Pectin is a fundamental component of the cell wall, and its presence confers rigidity and resistance to the fruit. Transient transformation experiments confirmed that increasing the expression of these genes enhances fruit firmness.
Therefore, this study lays an important foundation for understanding the genetic mechanisms involved, while also providing useful information for the genetic improvement of the Chinese cherry, opening up new possibilities for selecting varieties with harder and more resistant fruits.
In conclusion, the study of the Chinese cherry genome has provided important insights into the genetic basis of fruit firmness, offering new prospects for genetic improvement. Furthermore, understanding the molecular mechanisms underlying important quality traits, such as fruit texture, will enable breeders to develop Chinese cherry varieties with improved transport resistance.
Source: Jiu S, Lv Z, Liu M, Xu Y, Chen B, Dong X, Zhang X, Cao J, Manzoor MA, Xia M, Li F, Li H, Chen L, Zhang X, Wang S, Dong Y, Zhang C. (2024). Haplotype-resolved genome assembly for tetraploid Chinese cherry (Prunus pseudocerasus) offers insights into fruit firmness. Horticulture Research 8,11(7): uhae142. https://doi.org/10.1093/hr/uhae142.
Image: NAVER
Andrea Giovannini
University of Bologna (IT)
Cherry Times - All rights reserved











